Saturday, March 21, 2099
Tuesday, November 5, 2019
41) Running AVANA on a Mac / Linux machine
1) Download the tool executable from https://sourceforge.net/projects/avana/
2) Install the file by double clicking on the downloaded file. While installing, if you don't have certain java run time environment file, you will be promoted to download. Complete the download and installation of the Java files. Then try installing again. During the installation process, you would be asked where to install Avana. It would be preferred to not install this in the Applications folder, instead save it in an easy to find directory under your user profile, such "/Users/asif/Downloads/AVANA", where "AVANA" is a folder you created under the "Downloads".
3) Go to the folder where you installed AVANA. For this tutorial, let's say it is installed in "/Users/asif/Downloads/AVANA". Go to that folder and you should see a bunch of files, in particular a file entitled "runAvana.bat". We would need to either convert this Windows batch file (*.bat) to a Unix batch file (*.sh). To do this, either rename the windows batch file to "runAvana.sh" or create a new file with the name "runAvana.sh". You can choose to either keep or delete the "*.bat" file, it would not affect the execution of AVANA on a Mac / Linux machine.
4) Open the "runAvana.sh" file and delete the existing content inside. Then paste the following new content to the file:
#!/bin/bash
export AVANA_HOME=/Users/asif/Downloads/AVANA
export CLASSPATH=$AVANA_HOME/avana.jar:$AVANA_HOME/avana-gui.jar:$AVANA_HOME/lib/biojava-1.4.jar:$AVANA_HOME/lib/bytecode-0.92.jar:$AVANA_HOME/lib/commons-beanutils-1.7.0.jar:$AVANA_HOME/lib/commons-logging-1.1.jar:$AVANA_HOME/lib/commons-math-1.1.jar:$AVANA_HOME/lib/jcommon-1.0.0.jar:$AVANA_HOME/lib/jfreechart-1.0.1.jar
java -Xms256m -Xmx800m it.southdown.avana.ui.AvanaGUI
5) In your case, replace the path (/Users/asif/Downloads/AVANA) in "AVANA_HOME" line to where you have installed the tool.
6) Open the Terminal. Navigate to the folder (/Users/asif/Downloads/AVANA) on the Terminal.
7) Make the "runAvana.sh" file executable through the following command:
chmod a+x runAvana.sh
8) Execute Avana:
sudo ./runAvana.sh
9) Sudo is to give you admin access in case you need authorisation. Enter the administrator password of your computer when prompted.
10) Done! Enjoy measuring entropy!
Friday, August 27, 2010
40) How to include my own database ID along with the NCBI ID on the BLAST's result page?
>gi|34392270|emb|CAE46396.1| STAT_00044 Dd-STATb protein [Dictyostelium discoideum]
2. formatdb
formatdb -i input.fasta -p T -o T
3. In the /usr/local/wwwblast-2.2.17 folder of BioSLAX, backup blast.html and blast.cgi
4. Change the content of blast.cgi as shown (pipe the output of blast.REAL and do sed to get the hyperlink of your own database Id included in the blast result)
#!/bin/csh -f
#
# $Id: blast.cgi,v 1.1 2002/08/06 19:03:51 dondosha Exp $
#
echo "Content-type: text/html"
echo ""
#setenv DEBUG_COMMAND_LINE TRUE
setenv BLASTDB db
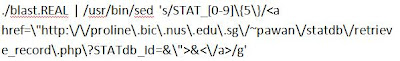
# I used my own database STATdb_Id as an example below.
5. Output example
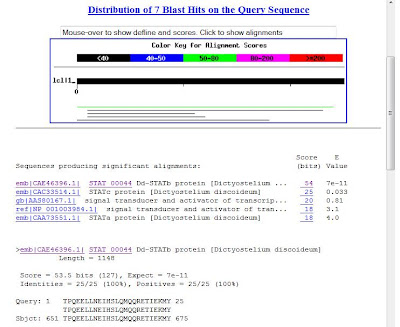
Thursday, March 11, 2010
Monday, March 8, 2010
Monday, December 14, 2009
37) LAPIS: Converting blast hit sequences into fasta format
For example, it can be used to extract only the subject and the identifier (NCBI GI number) information from a BLAST result. Below is an illustration of how this can be done using LAPIS:
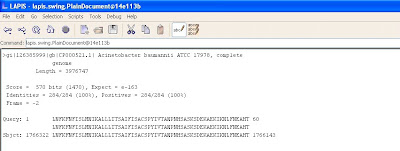
Before
>gi|126385999|gb|CP000521.1| Acinetobacter baumannii ATCC 17978, complete genome
Length = 3976747
Score = 570 bits (1470), Expect = e-163
Identities = 284/284 (100%), Positives = 284/284 (100%)
Frame = -2
Query: 1 LNFKFNFISLMNIKALLLITSAIFISACSPYIVTANPNHSASKSDEKAEKIKNLFNEAHT 60
LNFKFNFISLMNIKALLLITSAIFISACSPYIVTANPNHSASKSDEKAEKIKNLFNEAHT
Sbjct: 1766322 LNFKFNFISLMNIKALLLITSAIFISACSPYIVTANPNHSASKSDEKAEKIKNLFNEAHT 1766143
After
>gi|126385999
LNFKFNFISLMNIKALLLITSAIFISACSPYIVTANPNHSASKSDEKAEKIKNLFNEAHT
Methodology:
1. Select the line containing the fasta description together with the line containing the subject sequences: “line containing > or line containing sbjct” ->Tools ->Extract
2. To get rid of the numbers in the line containing sbjct: “digits in line containing sbjct” ->Tools -> Omit
3. To get rid of sbjct: “sbjct:” -> Extract -> Omit
4. To get rid of dashes: type “-" -> Extract -> Omit
5. To get rid of the extra spaces in the lines containing sequences: “spaces not in line containing >” -> Tools -> Omit
6. In case you want to clean up the description line to only have the GI
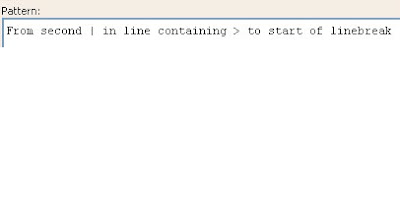
From second | in line containing > to start of linebreak
Screen shots: The following screen shots shows the input and the output at each step
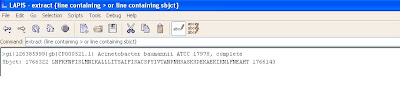
For extracting specific information, the user needs to find a pattern and type it in the pattern box as shown below:
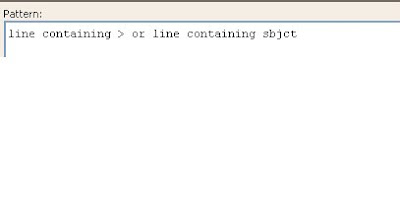
The pattern above is used to extract the two necessary lines for the further analysis.
Next the user should remove the positions (digits) in the subject line.
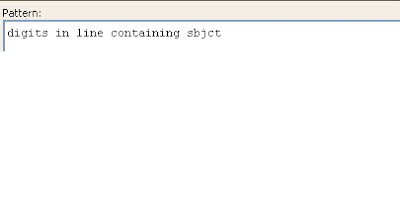
The screen shot below shows the highlighted digits to be omitted.
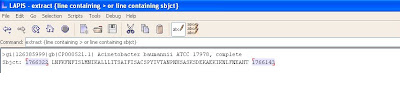
The screen shot below shows the information after omitting the numbers from the subject line.
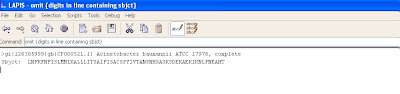
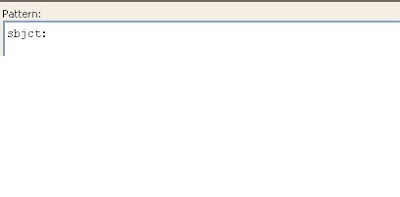
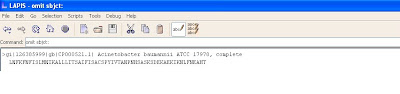
Next the word “subj:” should be omitted also.
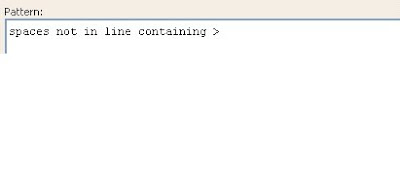
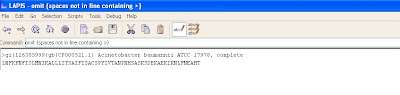
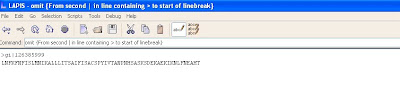
The screen shot below shows the pattern to remove any extra spaces in the subject line. In case of any gaps (-), they should also be removed.
The screen shot below shows the pattern to remove extra information from the header line.
The screen shot below shows the required output.
Sunday, September 27, 2009
36) OpenBabel GUI - file format converter
Basic operation:
OpenBabel converts chemical objects (currently molecules or reactions) from one file format to another. This interface is an alternative to a command line and has the same capabilities.
- Select the type of the type of the input file from the dropdown list.
- Click the "..." button and select the file. Its contents are displayed.
- Choose the output format and file in a similar way. You can merely display the output without saving it by not selecting an output file or by checking "Output below only..".
- Click the "Convert" button.
The message window below the button gives the number of molecules converted, and the contents of the output file are displayed.
By default, all the molecules in an input file are converted if the output format allows multiple molecules.
Saturday, September 26, 2009
35) Autodock 4.0 (Installation and working)
Autodock 4.0 can be found from the following website,
Kindly use the pdb and ligand files which comes along with the installation of Autodock 4.0, will be present in (MGLTools/MGL ToolsPckgs/Pmv/doc). Use hsg1.pdb (protein) and ind.pdb (ligand)Protein Preparation: File - Read Molecule - Open the file.pdb. Select - Select from string - Type HOH* (to remove water molecules)Edit - Add Polar only File - write PDB save ur file in pdbfile.pdb extension just without changing the path. just click and remove HETATM Click sort nodes before saving .
Ligand Preparation: Ligand - input open ur ligand.pdb - just note down the summary of the ligand. Torsion tree - choose root again go to torsion tree - detect root again go to torsion tree - show root expansion again go to torsion tree - choose torsions just click ok again go to torison tree - set number of torsions give the number of torisons that you get in the summary of the (TORSDOF) when u open the ligand. Output Save the ligand file in the same directory in ligandfile.pdbqt
Grid preparation: Select select from string just type your active site residues and then click ok Flexible residues - input macromolecule - choose macromolecule - click on your pdb from the panel displayed. Flexible residues - choose torsions from the currently selected residues - active site residues alone will be highlighted. Flexible residues - redisplay the macromolecule Flexible residues - save the files in both flexible as (pdbfile_flex.pdbqt) rigid as (pdbfile_rigid.pdbqt) .Grid - macromolecule - open- just open ur pdb_rigid.pdbqt file, ignore the warnings, if you have any metal ions in your proteinn , just open ur grid file and u can find (0.00) for ur metal ions, just find the valency for them from net and type maunally ther and again save it.you use this now. (if you dont hav metal ions), just ignore the warnings. set map types - choose ligand - click on ur ligand from the panel displayed output - save in .gpf edit gpf - dontt do anything just click ok for the time being Run - run autogrid just browse and locate ur autogrid.exe file , just browse and locate file .gpf (grid parameter file) and note that everything should be in the same directory, were you saved your pdb in protien preparation, dont change the path for your wish, just save were it is saving just put your autodock.exe and autogrid.exe in the same directory were ur files has been saved
Docking: Docking - Macromolecule - set rigid file name - just again open ur pdb_rigid.pdbqt file Ligand - choose ur ligand search parameters- just delete one zero from the 2500000 (just make it to 4 zeros), otherwise it wil take very long time. Docking parameters- default other option-autodock 4 parameters- default output - save in ligandfile.dpf Edit DPF - just leave and click ok Run - run autock just browse and locate ur autodock.exe file just browse and locate file .dpf (dock parameter file) and then click ok.
Analysing the results:open the widget "Analyse" and click open, locate the ligand.dlg file,Next click on to Macromolecule found below Dockings, just click open, it will open by default, if you cannot able to view your protein properly, simply press "N" button from the keyboard.
To view your results click on to conformations -play,
 It will open a widget shown in the picture.
It will open a widget shown in the picture.
click on to the ampersand (&) symbol.
 Analyse your results..!!
Analyse your results..!!
NOTE: kindly save all your files in the same folder where you have been installed your MGLTools and executable files, if you want to run the programme from your favourite folder of your choice, make sure that you place just the executable files alone in the place were you have saved your all files that you have been used during the course of working on Autodock, because without these executable files the programme wont run properly.
For more details regarding the algorithm,f orcefields and interpretation of the data kindly go through the Autodock manual which is available at below mentioned web site, http://autodock.scripps.edu/faqs-help/tutorial/using-autodock-4-with-autodocktools34) How to set up web blast using BioSlax
A guide to using BioSlax and/or Installing BioSlax as a full linux system to your hard drive
1. First, get your sequence dataset ready in FASTA format
2. Create a directory (for example: /root/Desktop/db) and save the FASTA file of interest to this newly created directory
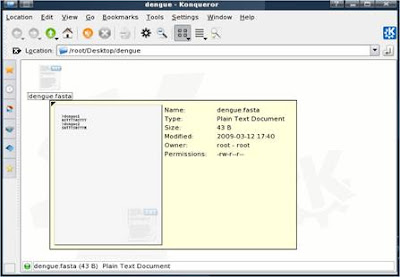
3. Now convert the FASTA formatted sequences into a blastable format, to do this, use the program “formatdb” which available for download from NCBI at http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download
*For BioSlax user, you do not need to download formatdb for BioSlax because it comes preinstalled
4. At console terminal, use this command to invoke Formatdb:
formatdb –i
Examples:
#formatdb –i ecoli.nt –p F
#formatdb -i swissprot -p T -o T
–p => type of sequence file;
T(rue) => the fasta file contains protein sequences
F(alse) => the fasta file contains nucleotide sequences
-o T => create an index file so that it is faster to run BLAST (optional)
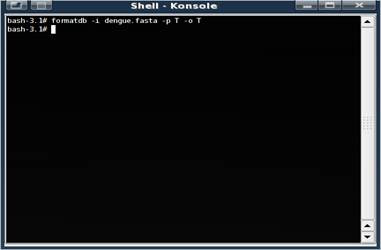
If the command is successful, a bunch of new files are created.
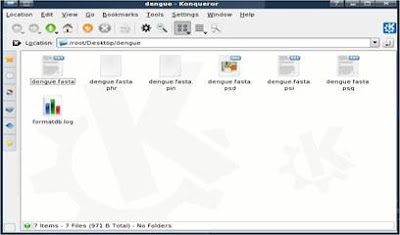
5. Now, go to K-menu => BioSLAX => Desktop Apps => BLAST DB setup
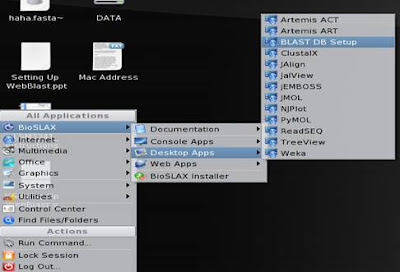
6. Fill up or browse the path of the directory containing the formatdb formatted sequence files (the directory you created at part 1) => click on “Set BLAST DB Now!” for Web Blast to do the necessary setup configurations
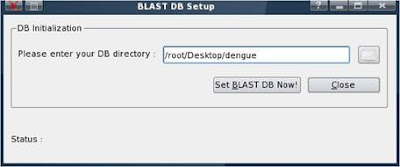
* Please note that the example “/root/Desktop/db” directory is fine if it is on a hard disk or a “configsave” has been done. If it's on a machine booted by LiveCD & no “configsave” is done, the files will be lost when the system is shutdown. Therefore, we recommend that user to create the database outside the virtual environment, i.e in a USB hdd or thumb drive.
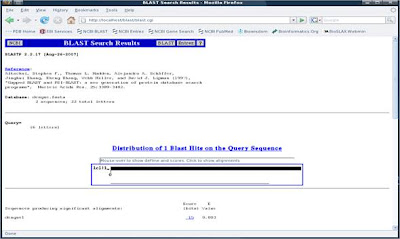
7. We are done, below are the results you will see:

BLAST result page
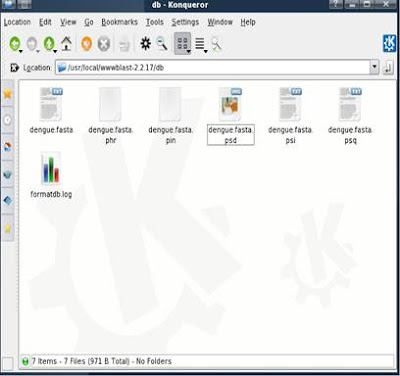
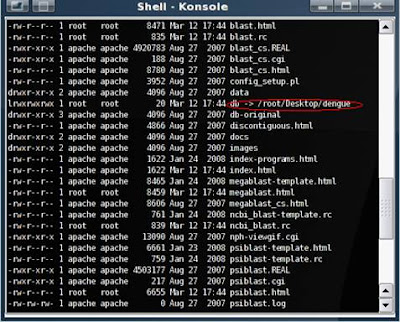
The location of your database after setting up BLAST Db: /usr/local/wwwblast-2.2.17/db
33) How to set-up the bioslax laptop servers
A guide to using BioSlax and/or Installing BioSlax as a full linux system to your hard drive
1. First of all, you need to change the password:
- Run the Konsole terminal
- bioslax#: passwd
- follow the instructions on the screen (please don’t set simple password)

2. To change host name: (skip this step if it has already been done)
- Run the Konsole terminal
- root@slax: pico /etc/HOSTNAME
- Change “bioslax.maxonline.com.sg” to your host+domain name (e.g.group01.bic.nus.edu.sg).
- Reboot
3. If the previous step 2 did not work, do the following, otherwise skip to step 4:
- Run the Konsole terminal
- bioslax#: cd /etc/rc.d
- bioslax#: pico rc.M
- You will be in the file rc.M. Type ctrl-W to search for "HOSTNAME". Type ctrl-W again, press enter and it will search the next word containing "HOSTNAME". You should be looking for the line "Set hostname by using DHCP response".
- Comment the whole paragraph from "set hostname by using DHCP response" to "done" by typing # at the start of each line.
- Save the changes by typing ctrl-X.
- Run the Konsole terminal again.
- bioslax#: netconfig
- There will be a blue window opened where you can enter the hostname, e.g."group01".
- Enter the domain name, e.g."bic.nus.edu.sg".
- Select "DHCP" as the network connection.
- At the "Set DHCP Hostname" window, leave it blank and press OK.
- Confirm the changes.
- Reboot
4. Set password for MYSQL
- Log in to Webmin (e.g. type http://group01.bic.nus.edu.sg:10000 for remote access) in Mozilla Firefox.
- Go to Servers
- Select MySQL Database Server
- Go to User Permissions
- Search for the row containing user “Anonymous”, host “localhost” and click on the "Anonymous" link to change the settings for that row:
Under “Permissions” Click “Select Table Data” (it will become highlighted) Click “Save” - Search for the row containing user “Anonymous”, host “bioslax” and click on the "Anonymous" link to change the settings for that row:
Under “Hosts” click the radio button next to the text-box and in the textbox type in “groupXX”
Under “Permissions” Click “Select Table Data” (it will become highlighted)
Click “Save” - Search for the row containing user “root”, host “localhost” and click on the "Anonymous" link to change the settings for that row:
Under “Password” click the radio button for “Set to” and fill in the root password in the text box
Click “Save” - Search for the row containing user “root”, host “bioslax” and click on the "Anonymous" link to change the settings for that row:
Under “Password” click the radio button for “Set to” and fill in the root password in the text box
Under “Hosts” click the radio button next to the text-box and in the textbox type in “groupXX”
Click “Save”
5. Three different ways to access the server remotely
- WinSCP (Freeware; http://winscp.net/).
- Hostname: group02.bic.nus.edu.sg
- Port number: 22
- Username: root (or any other username that you may have created on the server)
- Password: (type your password)
- Protocol: SFTP (allow SCP fallback)
- Click “Login”. If prompted for security alert, click “Yes”.
- Hostname: group02.bic.nus.edu.sg
- Putty (Freeware; http://www.putty.nl/).
- Hostname: group02.bic.nus.edu.sg
- Port number: 22
- Connection type: SSH
- Click “Open”. If prompted for security alert, click “Yes”.
- Login as: root (or any other user id)
- Password: (type your password)
- Hostname: group02.bic.nus.edu.sg
- Web browser (URL:http://group02.bic.nus.edu.sg)
- To access files from the web, you should save your web pages in the “/var/www/htdocs” folder
- To access pre-installed programs in Bioslax, such as blast and clustalw:
- To access files from the web, you should save your web pages in the “/var/www/htdocs” folder
Wednesday, September 2, 2009
32) Duplicates Finder - a java standalone application to find sequence duplicates in your dataset
One could use the ‘Remove duplicates’ function in Excel to do this; it returns the list of unique sequences. However, Excel does not do a good job when the dataset is large (large number or long sequences).
Here, we provide a simple desktop application to extract the sets of duplicates from a dataset and also provide the list of unique sequences from the input dataset.
How to use the program:
- Download the zip file DuplicateFinder.zip here. Extract contents to a desired location.
- Click on the DuplicatesFinder.jar, the program starts and you are good to go. Since this is a java program, it is OS independent.
(NOTE: You need to have java runtime environment (JRE) installed in your system to run the program. To check if your system has java installed, refer to the end of this post.)
Input Files accepted by the program:
- Comma separated values (*.csv)
- The data should contain two colums – Identifier number, Sequence.
Identifier is a unique number to identify the sequence.
Sequence is the set of sequences to be analyzed.
- The data should contain two colums – Identifier number, Sequence.
- FASTA (*.fasta) or Tab delimited (*.txt)
- The sequences should be in fasta format as shown below
>716349
uquiertyqyjej
quiejhieq
>837468
oiweirjtyiwj
- The sequences should be in fasta format as shown below
Output Files created by the program:
The output files are created in the same directory and folder as the input file.
Two output files are created:
- Duplicates sequences File: A file containing the sets of duplicates (
Duplicates.csv). The columns in the file are: - Duplicate Set – the index of groups of duplicates;
- Count – the index of a duplicate sequence in the set of duplicates it belongs to. So each series of numbers from 1 to n represents a set of sequence duplicates with a total of n duplicate sequences in that set;
- Sequence – the duplicate sequences in ascending order (for easy visual checking).
- Identifier – the number from the identifier column in the .csv file or the identifier number after '>' in the .fas and .txt files.
- Remarks – remarks if any.
- Unique sequences file: A file containing the unique sequences from the dataset in fasta format (
Unique.fas). The file includes one representative from each set of duplicates.
NOTE: In both output files, the sequences are arranged in ascending order of the sequence.
A Working Example

- Input file with 3 unique sequences and 2 groups of duplicates
(Group 1 – 1A and 1B; Group 2 – 2A, 2B and 2C) - Start the Duplicate Finder program. Click ‘Open’. A file chooser with file filtering set to show only *.csv, *.fas or *.txt files pops up. Select your input file (here, Sample.fas). Click ‘Open’.
- The path of your input file is set. Click ‘Generate results’. Wait for the confirmation window saying “Done” to indicate that your results have been generated. Click ‘OK’.
- Go to the folder containing your input file. You will find two new files created with names
Duplicates.csv and Unique.fas. In our case, our input file was Sample. So the results are SampleDuplicates.csv and SampleUnique.fas. - Output file 1: SampleDuplicates.csv
Duplicate column: index for groups of duplicates.
In our example, there were two groups of duplicates in the input file.
Count column: the index of a duplicate sequence in the group of duplicates it belongs to.
In our example, group 1 had two duplicate sequences – Duplicate1A and Duplicate1B – both of which are indicated in the Sequence column, followed by their identifier in the next column. Group 2 had three duplicate sequences – Duplicate2A, Duplicate2B and Duplicate2C – which are the three sequences indicated.
Sequence column: the duplicate sequences in ascending order (for easy visual checking).
Identifier column: the number from the identifier column in the .csv file or the identifier number after '>' in the .fas and .txt files - Output file 2: SampleUnique.fas
This file contains the set of unique sequences from the input dataset in fasta format. The set of unique sequences includes one representative from each set of duplicates, usually the first one in the set of duplicates.
In our example, we have 3 unique sequences – Unique1, Unique2 and Unique3 – which are present in the output file. In addition, the output also contains one representative from each set of duplicates – Duplicate1A from the first set and Duplicate2A from the second set.





To check if your system has JRE:
- Go to command prompt and type java –version.
- If your system has java, you will see the version details similar to one shown:
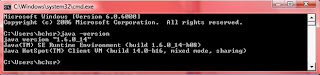
a) If your system does not have java installed, you get the error:
‘java’ is not recognized as an internal or external command, operable program or batch file.
b) You can then download the JRE for your OS from Sun Microsystems: http://www.java.com/en/download/manual.jsp)
Posted by: Rashmi
Edited by: Asif M. Khan
Thursday, August 20, 2009
31) Multiple sequence titles in the same BLAST hit
Are these sequences the same? And are they the same only at the aligned region or in the rest of the sequence as well?
I did a check on how these sequences are related. Based on several examples (two listed below), for now, it could be concluded that these entries that fall under the same BLAST hit are full sequence duplicates. However, in terms of annotations they could be from different isolates.
Case 1: 2 accession numbers under the same BLAST hit
 Alignment between full sequences of Q6R325.1 and AAS01728.1
Alignment between full sequences of Q6R325.1 and AAS01728.1- 100% identity, i.e. full sequences are exactly the same
- Same author
- Swissprot entry (Q6R325.1) has cross-reference to Genbank entry (AAS01728.1)
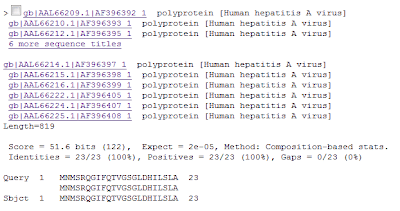 Multiple alignment of full sequences of all sequence titles
Multiple alignment of full sequences of all sequence titles- Perfect alignment
- Same author
- Only the isolate annotation is different
Wednesday, August 19, 2009
30) Using VLOOKUP (or HLOOKUP) in Microsoft Excel
- Data comparions between sheets
- Checking for discrepancies
- Linking data between sheets
The formula looks like this:
VLOOKUP(lookup_value,table_array,col_index_num,range_lookup)
--
lookup_value: The value to search in the first column of the table array
table_array: Two or more columns of data. The values in the first column of table_array are the values searched by lookup_value. These values can be text, numbers, or logical values. Uppercase and lowercase text is equivalent.
col_index_num: The column number in table_array from which the matching value must be returned. A col_index_num of 1 returns the value in the first column in table_array; a col_index_num of 2 returns the value in the second column in table_array, and so on. If col_index_num is:
- Less than 1, VLOOKUP returns the #VALUE! error value.
- Greater than the number of columns in table_array, VLOOKUP returns the #REF! error value.
range_lookup: A logical value that specifies whether you want VLOOKUP to find an exact match or an approximate match:
- If FALSE, VLOOKUP will only find an exact match. In this case, the values in the first column of table_array do not need to be sorted. If there are two or more values in the first column of table_array that match the lookup_value, the first value found is used. If an exact match is not found, the error value #N/A is returned.
- If TRUE or omitted, an exact or approximate match is returned. If an exact match is not found, the next largest value that is less than lookup_value is returned.
The values in the first column of table_array must be placed in ascending sort order; otherwise, VLOOKUP may not give the correct value. You can put the values in ascending order by choosing the Sort command from the Data menu and selecting Ascending. For more information, see Default sort orders.
A good reference for more details of the formula can be found here.
These details are illustrated in the following example.
Step 1:
a) Once you have your table_array (i.e. your “reference dataset” with the first column containing), and the list which you would like to lookup for, enter the VLOOKUP formula in the cell as follows:
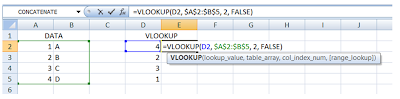
b) Note that in the formula, I have put in a “$” sign for the data source table indexes. This is to lock the table_array as A2:B5 and not allow it to slide as we drag the formula down to the rest of the cells.
This is what it’ll look like if I had not put the “$” sign there:
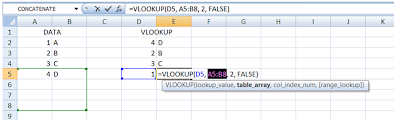
Notice that the data source table_array indexes have shifted (green box) and hence the value “1” could not be found. This will result in a #N/A error, as follows:
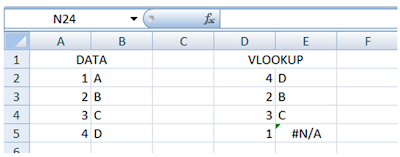
To avoid this, it is safer to use the “$” sign in your formula as shown in the Step 1(a).
Step 2: Hover on the edge of the cell, till your cursor becomes a “+”.
Click and drag the formula down to fill the rest of the cells.
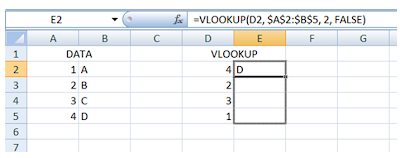
Step 3: And this is what you’ll get:
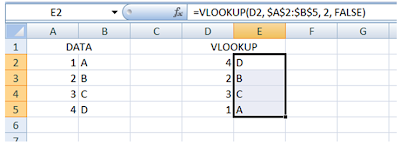
For data comparions from different sheets:
If your source/reference data is stored in another sheet, simply add the sheet name at the beginning of the [table_array] field, like this:
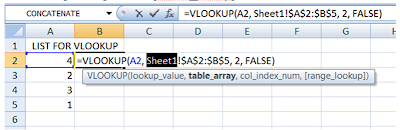
What if I have a horizontal set of data as my table_array?
The VLOOKUP formula described above is used when the table_array is vertical and the lookup values are in the first column.
Use HLOOKUP when the table_array is horizontal and the lookup values are in the top row. The other details, such as using the “$” sign, remain the same.
What does #N/A mean?
The error #N/A indicates that the source for a particular data was not found in the table_array. This could mean either (i) your table_array had shifted, as explained above in Step 1(b) or (ii) the data of interest is not present in the reference table.
Counting the #N/A's
Sometimes we might be interested in the number of data entries that do not have a match in the table_array (the reference table).
To count the number of #N/A errors in a dataset, use this formula:
=IF(ISNA(A1),1,0)
This prints 1 if cell "A1" contains #N/A, else prints 0. Sum up the column to obtain a total.




