1) Load your taln file (e.g. All_C.taln) in AVANA
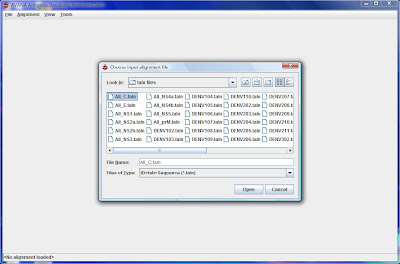
2) Load your metadata (e.g. All_C.csv) in AVANA
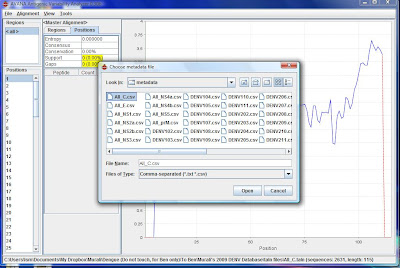
3) Go to Alignment\Create Subset
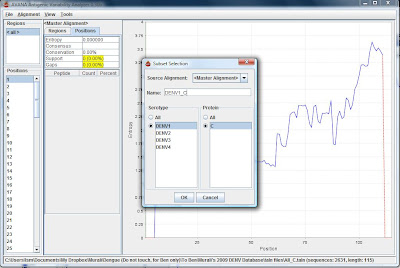
4) In the window, Source Alignment: Master Alignment, choose DENV1 as serotype and C as protein, and give a name to the subset (e.g. DENV1_C)
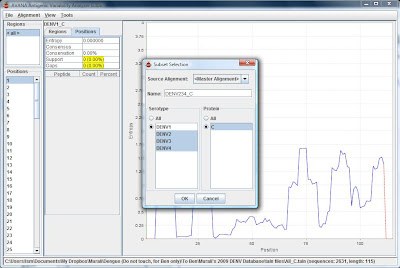
5) Create the next subset by repeating step 4, but this time highlight the rest (DENV2,DENV3 and DENV4) as serotype, and give another name to the subset (e.g.DENV234_C)
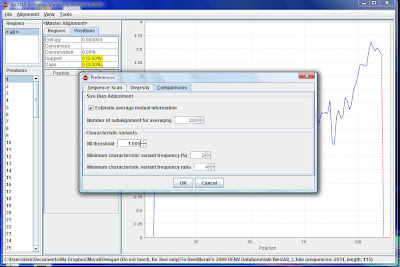
6) Check that the sample size is 9 by going to Tools\Preferences. It will scan for peptides of length 9 which is the typical length of t-cell epitopes.
7) In the same window, under the "Comparisons" tab, tick "Estimate average mutual information" and set the MI threshold to 1.000 (ideally we want value of 1, if get nothing then we consider other thresholds).
8) Next, go to View\Compare two alignments.
9) Click DENV1_C as top alignment and DENV234_C as bottom alignment
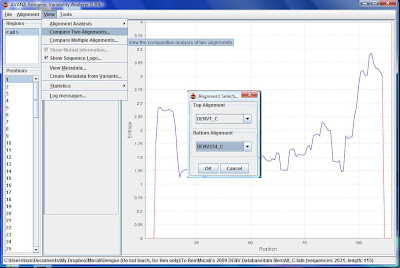
10) You will see two graphs. Go to View\Show Mutual Information and View\Show sequence logo.
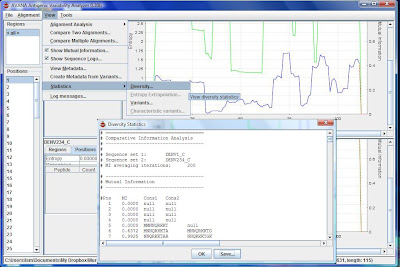
11) The green line in the graph shows the mutual information. MI value tells u what is characteristic in one alignment when compared to the other alignment. MI value of 1 means, it is seen in this alignment, but was not seen in the other alignment.
12) Click on the position numbers to your left to see the sequence logo, which shows conservation and all the peptides at that position.
13) Export your results by going to View\Statistics\Diversity and save as .txt. Create an excel sheet for better viewing. Sort them according to descending MI, then increasing entropy.



No comments:
Post a Comment