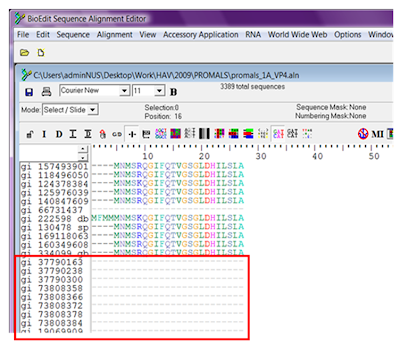
 We intend to remove sequences that are fully gapped, as shown in red box above.
We intend to remove sequences that are fully gapped, as shown in red box above.To do this,
- Click on: Sequence >> Filter out sequences containing certain characters >> Cut them to new alignment

- Select Contain non- and type “-“ in the textbox.
This means we are asking the tool to extract any sequence that contains a non-gap character.
- Save the output as a new .aln file, and you’ll have an alignment file without the fully gapped sequences!
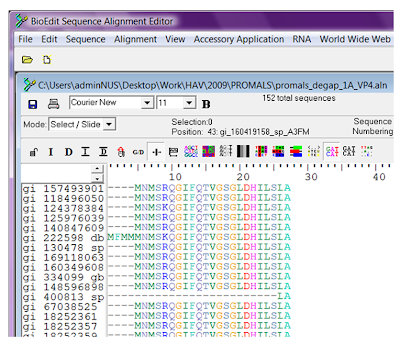
The sequences that are not fully gapped will be extracted to a new alignment, preserving the order of the sequences in the original alignment.



No comments:
Post a Comment