One could use the ‘Remove duplicates’ function in Excel to do this; it returns the list of unique sequences. However, Excel does not do a good job when the dataset is large (large number or long sequences).
Here, we provide a simple desktop application to extract the sets of duplicates from a dataset and also provide the list of unique sequences from the input dataset.
How to use the program:
- Download the zip file DuplicateFinder.zip here. Extract contents to a desired location.
- Click on the DuplicatesFinder.jar, the program starts and you are good to go. Since this is a java program, it is OS independent.
(NOTE: You need to have java runtime environment (JRE) installed in your system to run the program. To check if your system has java installed, refer to the end of this post.)
Input Files accepted by the program:
- Comma separated values (*.csv)
- The data should contain two colums – Identifier number, Sequence.
Identifier is a unique number to identify the sequence.
Sequence is the set of sequences to be analyzed.
- The data should contain two colums – Identifier number, Sequence.
- FASTA (*.fasta) or Tab delimited (*.txt)
- The sequences should be in fasta format as shown below
>716349
uquiertyqyjej
quiejhieq
>837468
oiweirjtyiwj
- The sequences should be in fasta format as shown below
Output Files created by the program:
The output files are created in the same directory and folder as the input file.
Two output files are created:
- Duplicates sequences File: A file containing the sets of duplicates (
Duplicates.csv). The columns in the file are: - Duplicate Set – the index of groups of duplicates;
- Count – the index of a duplicate sequence in the set of duplicates it belongs to. So each series of numbers from 1 to n represents a set of sequence duplicates with a total of n duplicate sequences in that set;
- Sequence – the duplicate sequences in ascending order (for easy visual checking).
- Identifier – the number from the identifier column in the .csv file or the identifier number after '>' in the .fas and .txt files.
- Remarks – remarks if any.
- Unique sequences file: A file containing the unique sequences from the dataset in fasta format (
Unique.fas). The file includes one representative from each set of duplicates.
NOTE: In both output files, the sequences are arranged in ascending order of the sequence.
A Working Example

- Input file with 3 unique sequences and 2 groups of duplicates
(Group 1 – 1A and 1B; Group 2 – 2A, 2B and 2C) - Start the Duplicate Finder program. Click ‘Open’. A file chooser with file filtering set to show only *.csv, *.fas or *.txt files pops up. Select your input file (here, Sample.fas). Click ‘Open’.
- The path of your input file is set. Click ‘Generate results’. Wait for the confirmation window saying “Done” to indicate that your results have been generated. Click ‘OK’.
- Go to the folder containing your input file. You will find two new files created with names
Duplicates.csv and Unique.fas. In our case, our input file was Sample. So the results are SampleDuplicates.csv and SampleUnique.fas. - Output file 1: SampleDuplicates.csv
Duplicate column: index for groups of duplicates.
In our example, there were two groups of duplicates in the input file.
Count column: the index of a duplicate sequence in the group of duplicates it belongs to.
In our example, group 1 had two duplicate sequences – Duplicate1A and Duplicate1B – both of which are indicated in the Sequence column, followed by their identifier in the next column. Group 2 had three duplicate sequences – Duplicate2A, Duplicate2B and Duplicate2C – which are the three sequences indicated.
Sequence column: the duplicate sequences in ascending order (for easy visual checking).
Identifier column: the number from the identifier column in the .csv file or the identifier number after '>' in the .fas and .txt files - Output file 2: SampleUnique.fas
This file contains the set of unique sequences from the input dataset in fasta format. The set of unique sequences includes one representative from each set of duplicates, usually the first one in the set of duplicates.
In our example, we have 3 unique sequences – Unique1, Unique2 and Unique3 – which are present in the output file. In addition, the output also contains one representative from each set of duplicates – Duplicate1A from the first set and Duplicate2A from the second set.





To check if your system has JRE:
- Go to command prompt and type java –version.
- If your system has java, you will see the version details similar to one shown:
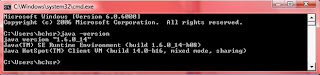
a) If your system does not have java installed, you get the error:
‘java’ is not recognized as an internal or external command, operable program or batch file.
b) You can then download the JRE for your OS from Sun Microsystems: http://www.java.com/en/download/manual.jsp)
=UPDATE=
This program is only able to remove exact EQUAL LENGTH duplicate sequences; it does not treat subset duplicates as duplicates. For example, let's say we have the following sequences:
>1
AAAAAAAAA
>2
AAAAAAAA
>3
AAAAAAAA
The output of unique sequences from the program are:
>1
AAAAAAAAA
>2
AAAAAAAA
You can see that though >2 is a subset duplicate of >1, the program still considers it as unique because it only removes exact same length duplicate sequences, such as >2 and >3, where one was removed and the other kept.
What happens if you have gaps in your input sequences? Let's say we have the following input sequences:
>1
AAAAAAAAA
>2
AAAAAAAA-
>3
AAAAAAAA-
>4
AAAAAAAAAB
>5
AAAAAAAAAB
>6
AAAAAAAAAC
The output that you get from the program is:
>2
AAAAAAAA-
>1
AAAAAAAAA
>4
AAAAAAAAAB
>6
AAAAAAAAAC
Just like in the previous example, only exact equal length duplicates are removed and the gap is treated like an amino acid. So, if you do not want gaps to affect the removal of duplicates (especially if they are in the middle of the sequences), you should remove them using the simple replace function in most text editors (replace "-" with nothing).
So what if you are interested in removing subset duplicates? What program can you use?
You can use Jalview and a tutorial is available here.
Content by: Rashmi & Asif M. Khan
Posted by: Rashmi
Edited by: Asif M. Khan
Posted by: Rashmi
Edited by: Asif M. Khan



Hi,
ReplyDeleteI have been looking for something like this for a while. Thank you so much making this! I previously used Jalview's feature to remove duplicates, but for some reasons the results were not consistent. It was frustrating..you are my saviour! Thank you again
http://proline.bic.nus.edu.sg/~asif/tools/DuplicateFinder.zip appears to be not accessible.
ReplyDeleteSorry, server is down till Monday, some maintenance work going on..thx for your patience.
ReplyDeleteThanks, the link is alive.
ReplyDeletewhat about if you have a million reads like from NGS? Is there anyone who tried it?
ReplyDeletegood luck
DeleteI am unable to open the tool in java. can any one help me out...
ReplyDelete@Anonymous: No we haven't tried such large datasets.
ReplyDelete@Ghana: May I know what do you mean by open the tool in java? You just need to extract files from the zip folder. In the extracted folder, you will see a folder called lib and a jar file DuplicatesFinder v1.1.jar. Just double click this file to use the tool.
Thank you very much for making such a wonderful software. I had a big file with 3000 duplicates. I spent more two days in removing 3000 duplicates. but this software did the job in seconds.
ReplyDeleteAre you working on making another good software also.
thanks
Is the http://proline.bic.nus.edu.sg/~asif/tools/DuplicateFinder.zip currently out of order?
ReplyDeleteHi, great software! Very useful and I appreciate your work.
ReplyDeleteI do have one question though.
The output file "Duplicates sequences" didn´t create columns for the five headers so that "Duplicate Set,Count,Sequence,Identifier,Remarks"
are all given in one line which makes it difficult to summarize.
Am I doing something wrong?
Thanks!
Just FIY, In the current zip file, the lib directory isn't present for some reason. Program works if you create it and put all the files (Except duplicatefinder.jar) into it.
ReplyDeleteThis was very useful, thank you!
ReplyDelete