>gi|34392270|emb|CAE46396.1| STAT_00044 Dd-STATb protein [Dictyostelium discoideum]
2. formatdb
formatdb -i input.fasta -p T -o T
3. In the /usr/local/wwwblast-2.2.17 folder of BioSLAX, backup blast.html and blast.cgi
4. Change the content of blast.cgi as shown (pipe the output of blast.REAL and do sed to get the hyperlink of your own database Id included in the blast result)
#!/bin/csh -f
#
# $Id: blast.cgi,v 1.1 2002/08/06 19:03:51 dondosha Exp $
#
echo "Content-type: text/html"
echo ""
#setenv DEBUG_COMMAND_LINE TRUE
setenv BLASTDB db
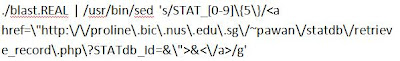
# I used my own database STATdb_Id as an example below.
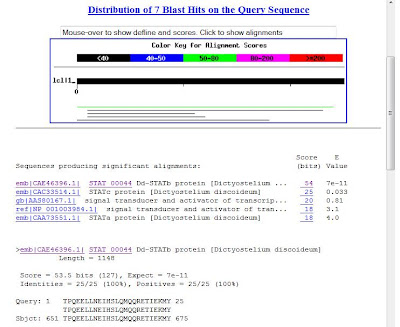
5. Output example



what do the parameter of blast.REAL mean?
ReplyDeleteHow could we fix for our own website?
is it only for c shell?
It looks nice, but you did not precise what it the script for retrieve_record.php. Can you tell us?
ReplyDelete