For a more comprehensive and updated information on this, please see Post 32.
Read all the way to the end of that post.
======================
You can use Jalview to easily check for duplicates and remove them if any.
1. Download and install Jalview on your home system from
http://www.jalview.org/download.html

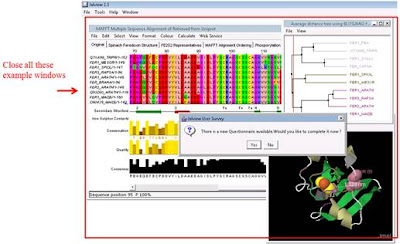
2. Run Jalview and close all example windows
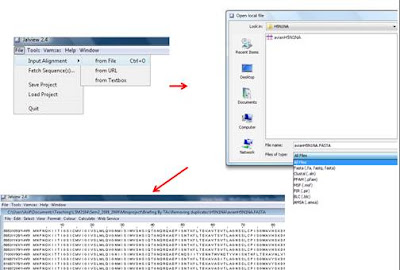
3. Load your fasta file to Jalview
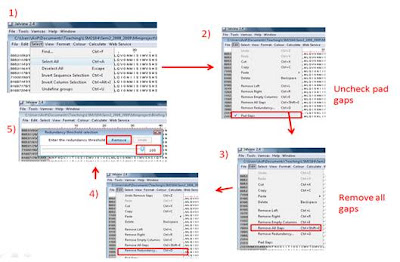
4. Remove duplicates:
- Select all sequence
- Go ‘Edit’ to uncheck the pad gaps function.
- In ‘Edit’, select ‘Remove all gaps’
- After that select ‘Remove redundancy’
- At the “redundancy threshold selection” dialog box, set the threshold value to 100, click ‘Remove’.
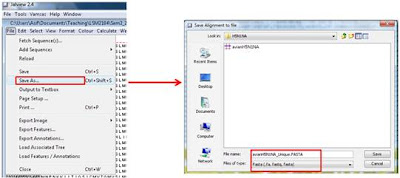
5. Saving the unique fasta file and you are done!
Content by: Asif M. Khan & Sye Bee
Posted by: Sye Bee
Edited by: Asif M. Khan



great, I like it!
ReplyDeleteThank ya for the help
ReplyDeleteHere is my free program on Github **Sequence database curator**
ReplyDelete(https://github.com/Eslam-Samir-Ragab/Sequence-database-curator)
It is a very fast program and it can deal with:
1. Nucleotide sequences
2. Protein sequences
It can work under Operating systems:
1. Windows
2. Mac
3. Linux
It also works for:
1. Fasta format
2. Fastq format
Best Regards