A guide to using BioSlax and/or Installing BioSlax as a full linux system to your hard drive
1. First, get your sequence dataset ready in FASTA format
2. Create a directory (for example: /root/Desktop/db) and save the FASTA file of interest to this newly created directory
3. Now convert the FASTA formatted sequences into a blastable format, to do this, use the program “formatdb” which available for download from NCBI at http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download
*For BioSlax user, you do not need to download formatdb for BioSlax because it comes preinstalled
4. At console terminal, use this command to invoke Formatdb:
formatdb –i
Examples:
#formatdb –i ecoli.nt –p F
#formatdb -i swissprot -p T -o T
–p => type of sequence file;
T(rue) => the fasta file contains protein sequences
F(alse) => the fasta file contains nucleotide sequences
-o T => create an index file so that it is faster to run BLAST (optional)
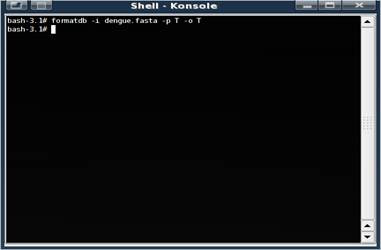
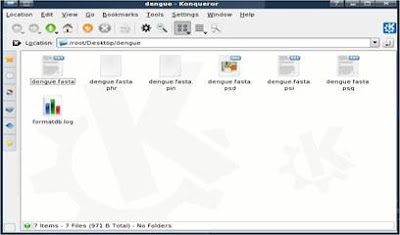
If the command is successful, a bunch of new files are created.
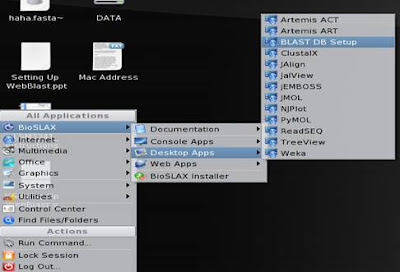
5. Now, go to K-menu => BioSLAX => Desktop Apps => BLAST DB setup
6. Fill up or browse the path of the directory containing the formatdb formatted sequence files (the directory you created at part 1) => click on “Set BLAST DB Now!” for Web Blast to do the necessary setup configurations
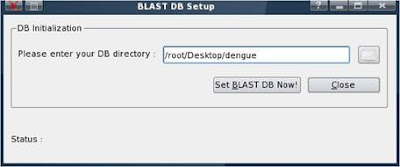
* Please note that the example “/root/Desktop/db” directory is fine if it is on a hard disk or a “configsave” has been done. If it's on a machine booted by LiveCD & no “configsave” is done, the files will be lost when the system is shutdown. Therefore, we recommend that user to create the database outside the virtual environment, i.e in a USB hdd or thumb drive.
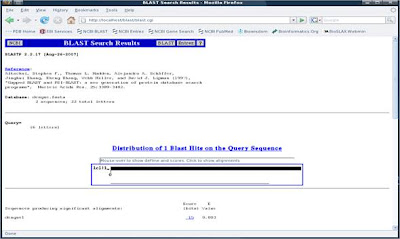
7. We are done, below are the results you will see:

BLAST result page
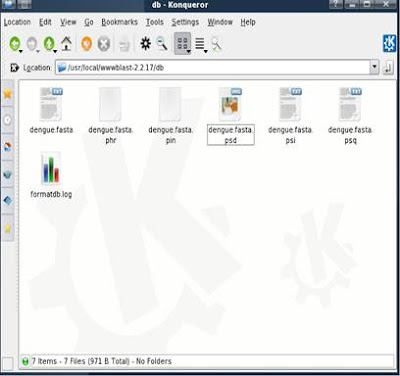
The location of your database after setting up BLAST Db: /usr/local/wwwblast-2.2.17/db



No comments:
Post a Comment